Data Technology Notes
Data Technologies
Class and homework examples often start with a nice, clean, rectangular data set.
Once you have a source for your data, data technologies are the tools you need to get your data to that point.
The process of applying these tools is sometimes called data wrangling.
The main steps are usually
importing the data;
cleaning the data;
merging information from several sources;
transforming to specialized data types
reshaping the data.
We have seen many of these steps already.
Data Import
Your data may be available in a number of different forms, such as:
as a CSV file;
another form of text file;
a spread sheet;
a table on a web page;
from a web service;
in a SQL data base
in some other type of data base or file format.
We have seen several examples of importing CSV files.
CSV files are often fairly easy to deal with.
They are typically meant for data import rather than human reading.
Spread sheet programs typically provide a mechanism for exporting data in CSV form.
Some issues that you need to look for:
identifying column names;
identifing row names;
use of a semicolon separator (common in Europe).
Unemployment data from the Local Area Unemployment Statistics (LAUS) page ofthe Bureau of Labor Statistics is an example of a structured text file not in CSV format.
These files usually have enough structure for automated import.
Often they have features designed for a human reader that need to be worked around.
Spread sheets are in principle designed for machine computation.
Many spread sheets have features designed for a human reader that need to be worked around.
Exporting (the relevant part of) a spread sheet to CSV is sometimes a good option.
Using a tool that can import directly from the spread sheet is often better: it makes the analysis easier to reproduce and repeat with new data.
Wrangling the Unemployment Data
The unemployment data is available as a text file.
The fields are delimited with a
|character and surrounded by white space.There is a header and a footer that need to be removed.
The counts use a comma to separate thousands.
The column names are awkward.
Some names contain an apostrophe, which can be confused with a quote character.
The base function read.table and the readr function read_table have many options for dealing with these issues.
We can use read.table with:
col.namesto specify column names;skip = 6to remove the header;strip_whiteto remove white space around itemsquote = ""to turn off interpreting any character as delimiting a quoted stringfill = TRUEto fill in columns in the footer.
lausURL <- "https://stat.uiowa.edu/~luke/data/laus/laucntycur14-2017.txt"
lausFile <- "laucntycur14-2017.txt"
if (! file.exists(lausFile))
download.file(lausURL, lausFile)
lausUS <- read.table(lausFile,
col.names = c("LAUSAreaCode", "State", "County",
"Title", "Period",
"LaborForce", "Employed",
"Unemployed", "UnempRate"),
quote = "", sep = "|", skip = 6, strip.white = TRUE,
fill = TRUE)
head(lausUS)
## LAUSAreaCode State County Title Period LaborForce Employed
## 1 CN0100100000000 1 1 Autauga County, AL Nov-16 25,809 24,518
## 2 CN0100300000000 1 3 Baldwin County, AL Nov-16 89,507 84,817
## 3 CN0100500000000 1 5 Barbour County, AL Nov-16 8,213 7,546
## 4 CN0100700000000 1 7 Bibb County, AL Nov-16 8,645 8,105
## 5 CN0100900000000 1 9 Blount County, AL Nov-16 24,754 23,481
## 6 CN0101100000000 1 11 Bullock County, AL Nov-16 4,990 4,609
## Unemployed UnempRate
## 1 1,291 5.0
## 2 4,690 5.2
## 3 667 8.1
## 4 540 6.2
## 5 1,273 5.1
## 6 381 7.6
tail(lausUS)
## LAUSAreaCode
## 45066 CN7215300000000
## 45067 --------------------------------------------------------------------------------------------------------------------------------------
## 45068 (p) = preliminary.
## 45069 Dash indicates that data are not available.
## 45070 SOURCE: BLS, LAUS
## 45071 February 6, 2018
## State County Title Period LaborForce Employed Unemployed
## 45066 72 153 Yauco Municipio, PR Dec-17(p) 10,523 8,892 1,631
## 45067 NA NA
## 45068 NA NA
## 45069 NA NA
## 45070 NA NA
## 45071 NA NA
## UnempRate
## 45066 15.5
## 45067
## 45068
## 45069
## 45070
## 45071Searching for a sequence of dashes allows the code to drop the footer to work even if the number of rows changes.
footstart <- grep("------", lausUS$LAUSAreaCode)
lausUS <- lausUS[1:(footstart - 1), ]str show the data types we now have:
str(lausUS)
## 'data.frame': 45066 obs. of 9 variables:
## $ LAUSAreaCode: chr "CN0100100000000" "CN0100300000000" "CN0100500000000" "CN0100700000000" ...
## $ State : int 1 1 1 1 1 1 1 1 1 1 ...
## $ County : int 1 3 5 7 9 11 13 15 17 19 ...
## $ Title : chr "Autauga County, AL" "Baldwin County, AL" "Barbour County, AL" "Bibb County, AL" ...
## $ Period : chr "Nov-16" "Nov-16" "Nov-16" "Nov-16" ...
## $ LaborForce : chr "25,809" "89,507" "8,213" "8,645" ...
## $ Employed : chr "24,518" "84,817" "7,546" "8,105" ...
## $ Unemployed : chr "1,291" "4,690" "667" "540" ...
## $ UnempRate : chr "5.0" "5.2" "8.1" "6.2" ...Convert the counts to numbers by removing the commas and passing to as.numeric:
lausUS <- mutate(lausUS,
LaborForce = as.numeric(gsub(",", "", LaborForce)),
Employed = as.numeric(gsub(",", "", Employed)),
Unemployed = as.numeric(gsub(",", "", Unemployed)))
## Warning: There were 3 warnings in `mutate()`.
## The first warning was:
## ℹ In argument: `LaborForce = as.numeric(gsub(",", "", LaborForce))`.
## Caused by warning:
## ! NAs introduced by coercion
## ℹ Run `dplyr::last_dplyr_warnings()` to see the 2 remaining warnings.The expression
gsub(",", "", x)replaces all occurrences of the pattern "," in the elements of x with the empty string "". Using sub instead of gsub would replace only the first occurrence in each element.
We could also separate out the state character codes and county names:
lausUS <- mutate(lausUS,
StateCharCode = sub("^.*, ", "", Title),
CountyName = sub(", .*", "", Title))We could also convert the Period variable to a date format.
Refugee Arrival Data
The Refugee Processing Center provides information from the United States Refugee Admissions Program (USRAP), including arrivals by state and nationality.
Three files are available locally:
data from early January 2017;
data from early April 2017.
data from early March 2018.
These are Excel spread sheets.
The function read_excel in the readxl package provides a way to import this kind of data into R.
Load the readxl package and define a short name for the file:
library(readxl)
fname <- "Arrivals-2017-01-06.xls"Read the FY line:
read_excel(fname, skip = 13, n_max = 1)
## # A tibble: 0 × 1
## # ℹ 1 variable: FY 2017 <lgl>
year_line <- read_excel(fname, skip = 13,
col_names = FALSE, n_max = 1)
## New names:
## • `` -> `...1`
as.numeric(sub("FY ", "", year_line[[1]]))
## [1] 2017A useful sanity check on the format might be:
stopifnot(length(year_line) == 1 && grepl("FY [[:digit:]]+", year_line[[1]]))
year <- as.numeric(sub("FY ", "", year_line[[1]]))Read in the data:
d <- read_excel(fname, skip = 16)
## New names:
## • `` -> `...1`
head(d)
## # A tibble: 6 × 18
## ...1 Nationality Oct Nov Dec Jan Feb Mar Apr May Jun Jul
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alaba… <NA> 8 4 4 0 0 0 0 0 0 0
## 2 <NA> Dem. Rep. … 4 0 0 0 0 0 0 0 0 0
## 3 <NA> Iraq 0 0 4 0 0 0 0 0 0 0
## 4 <NA> Somalia 4 4 0 0 0 0 0 0 0 0
## 5 Alaska <NA> 17 3 2 0 0 0 0 0 0 0
## 6 <NA> Republic o… 3 0 0 0 0 0 0 0 0 0
## # ℹ 6 more variables: Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>
tail(d)
## # A tibble: 6 × 18
## ...1 Nationality Oct Nov Dec Jan Feb Mar Apr May Jun Jul
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 <NA> Sudan 0 0 3 0 0 0 0 0 0 0
## 2 <NA> Syria 14 4 5 0 0 0 0 0 0 0
## 3 <NA> Thailand 0 0 1 0 0 0 0 0 0 0
## 4 <NA> Vietnam 7 0 0 0 0 0 0 0 0 0
## 5 Total <NA> 9945 8355 7371 0 0 0 0 0 0 0
## 6 Pleas… <NA> NA NA NA NA NA NA NA NA NA NA
## # ℹ 6 more variables: Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>The last line needs to be dropped. Another sanity chack first is a good idea.
stopifnot(all(is.na(tail(d, 1)[, -1])))
d <- d[seq_len(nrow(d) - 1), ]state_lines <- which(! is.na(d[[1]]))
d[state_lines, ]
## # A tibble: 50 × 18
## ...1 Nationality Oct Nov Dec Jan Feb Mar Apr May Jun Jul
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alab… <NA> 8 4 4 0 0 0 0 0 0 0
## 2 Alas… <NA> 17 3 2 0 0 0 0 0 0 0
## 3 Ariz… <NA> 547 493 310 0 0 0 0 0 0 0
## 4 Arka… <NA> 0 0 14 0 0 0 0 0 0 0
## 5 Cali… <NA> 814 887 636 0 0 0 0 0 0 0
## 6 Colo… <NA> 192 217 180 0 0 0 0 0 0 0
## 7 Conn… <NA> 47 73 63 0 0 0 0 0 0 0
## 8 Dist… <NA> 2 0 0 0 0 0 0 0 0 0
## 9 Flor… <NA> 267 265 276 0 0 0 0 0 0 0
## 10 Geor… <NA> 362 304 331 0 0 0 0 0 0 0
## # ℹ 40 more rows
## # ℹ 6 more variables: Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>We can use the first column as the destination:
library(dplyr)
d <- rename(d, Dest = 1)We need to replace the NA values in Dest by the previous non-NA value. There are several options:
- use a
forloop; - calculate the gaps and use
rep; - use the
fillfunction fromtidyr.
A for loop solution:
v0 <- d[[1]]
for (i in seq_along(v0)) {
if (! is.na(v0[i]))
s <- v0[i]
else
v0[i] <- s
}A solution using rep:
v1 <- rep(d[state_lines[-length(state_lines)], 1][[1]], diff(state_lines))A search on R carry forward brings up the fill function in tidyr:
v2 <- fill(d, Dest)[[1]]All three approaches produce identical results:
identical(v0, v1)
## [1] FALSE
identical(v0, v2)
## [1] TRUEUsing fill is easiest:
d <- fill(d, Dest)Dropping the state lines gives the date we need:
head(d[-state_lines, ])
## # A tibble: 6 × 18
## Dest Nationality Oct Nov Dec Jan Feb Mar Apr May Jun Jul
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alaba… Dem. Rep. … 4 0 0 0 0 0 0 0 0 0
## 2 Alaba… Iraq 0 0 4 0 0 0 0 0 0 0
## 3 Alaba… Somalia 4 4 0 0 0 0 0 0 0 0
## 4 Alaska Republic o… 3 0 0 0 0 0 0 0 0 0
## 5 Alaska Somalia 4 0 2 0 0 0 0 0 0 0
## 6 Alaska Ukraine 10 3 0 0 0 0 0 0 0 0
## # ℹ 6 more variables: Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>To be able to read new files from this source it is useful to wrap this in a function:
library(readxl)
library(dplyr)
library(tidyr)
readRefXLS <- function(fname) {
read_excel(fname, skip = 13, n_max = 1)
## read and check the FY line
year_line <- read_excel(fname, skip = 13,
col_names = FALSE, n_max = 1)
stopifnot(length(year_line) == 1 &&
grepl("FY [[:digit:]]+", year_line[[1]]))
year <- as.numeric(sub("FY ", "", year_line[[1]]))
d <- read_excel(fname, skip = 16)
## check and trim the last line
stopifnot(all(is.na(tail(d, 1)[, -1])))
d <- d[seq_len(nrow(d) - 1), ]
## identify the state summary lines
state_lines <- which(! is.na(d[[1]]))
## rename and fill first column
d <- rename(d, Dest = 1)
d <- fill(d, Dest)
## drop the state summaries and add the FY
d <- d[-state_lines, ]
d$FY <- year
d
}This can read all the files:
d1 <- readRefXLS("Arrivals-2017-01-06.xls")
## New names:
## New names:
## • `` -> `...1`
d2 <- readRefXLS("Arrivals-2017-04-05.xls")
## New names:
## New names:
## • `` -> `...1`
d3 <- readRefXLS("Arrivals-2018-03-05.xls")
## New names:
## New names:
## • `` -> `...1`Some explorations:
td3 <- gather(d3, month, count, 3 : 14)
arr_by_dest <- summarize(group_by(td3, Dest), count = sum(count))
arr_by_nat <- summarize(group_by(td3, Nationality), count = sum(count))
arr_by_dest_nat <- summarize(group_by(td3, Dest, Nationality),
count = sum(count))
## `summarise()` has grouped output by 'Dest'. You can override using the
## `.groups` argument.ggplot(arr_by_dest, aes(count, Dest)) + geom_point()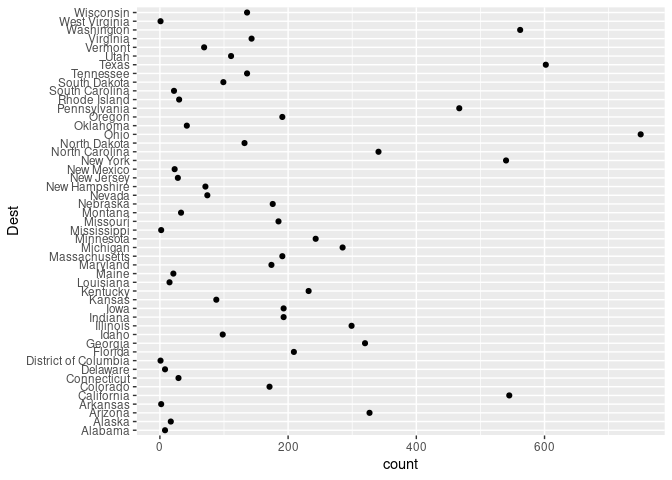
ggplot(arr_by_dest, aes(count, reorder(Dest, count))) + geom_point()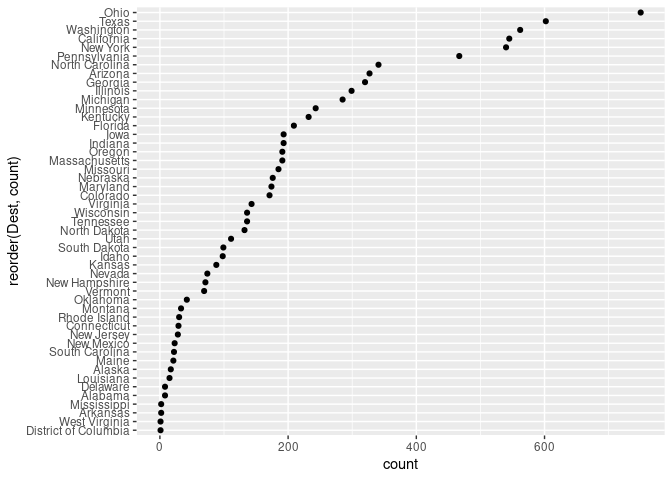
ggplot(slice_max(arr_by_dest, count, n = 5), aes(count, Dest)) + geom_point()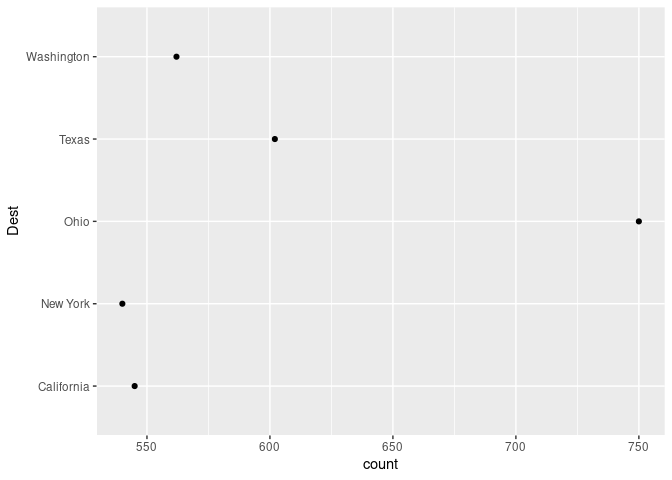
ggplot(arr_by_nat, aes(count, Nationality)) + geom_point()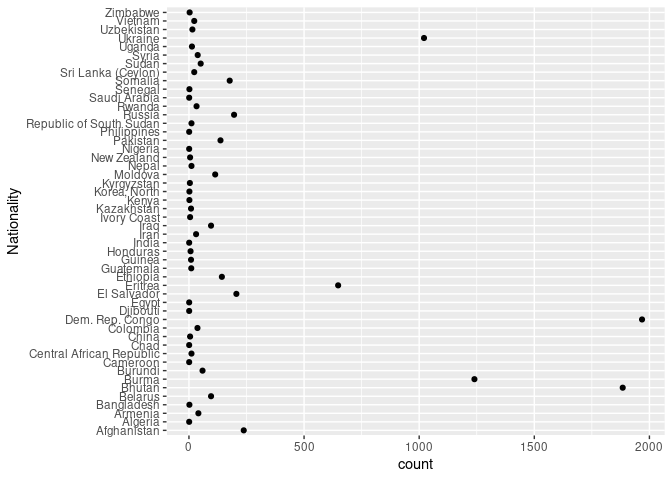
ggplot(arr_by_nat, aes(count, reorder(Nationality, count))) + geom_point()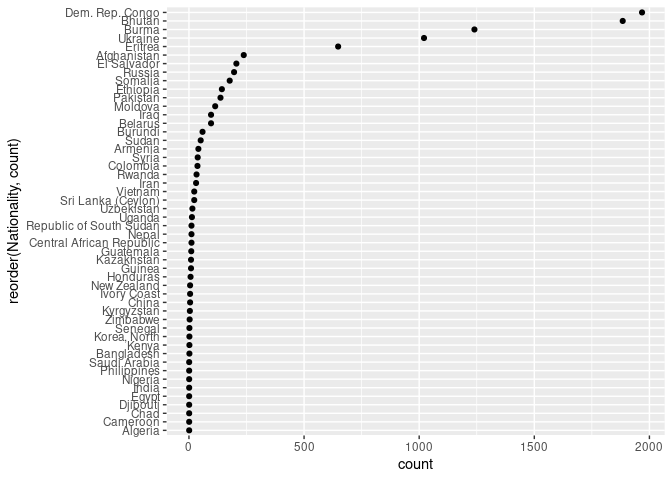
ggplot(slice_max(arr_by_nat, count, n = 5), aes(count, Nationality)) +
geom_point()
A useful visualization of flows is a Sankey diagram. These are also known as
river digrams;
and some would argue that allucial diagrams and river diagrams are different things.
Several R packages create these; including
alluvialriverplotgoogleVis.
This code creates a Sankey diagram using alluvial:
n_top <- 10
s_top <- 25
toprefs <- arrange(arr_by_nat, desc(count))$Nationality[1 : n_top]
topstates <- arrange(arr_by_dest, desc(count))$Dest[1 : s_top]
td3m <- mutate(td3,
Nationality = ifelse(Nationality %in% toprefs,
Nationality, "Other Nat."),
Dest = ifelse(Dest %in% topstates, Dest, "Other Dest."))
std3m <- summarize(group_by(td3m, Dest, Nationality), count = sum(count))
## `summarise()` has grouped output by 'Dest'. You can override using the
## `.groups` argument.
pal <- c(rainbow(n_top), "grey")
pal <- c(RColorBrewer::brewer.pal(n_top, "Paired"), "grey")
src <- c(toprefs, "Other Nat.")
library(alluvial)
with(std3m, alluvial(Nationality, Dest, freq = count,
col = pal[match(Nationality, src)],
cex = 0.8, alpha = 0.7))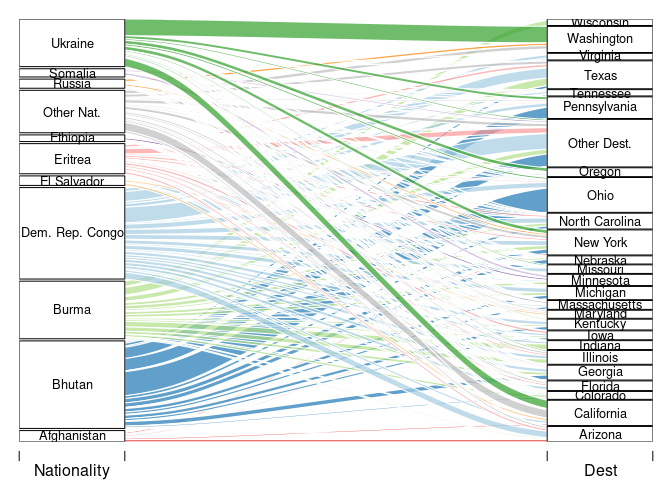
Another option is a chord diagram:
library(circlize)
chordDiagram(select(std3m, Nationality, Dest, count))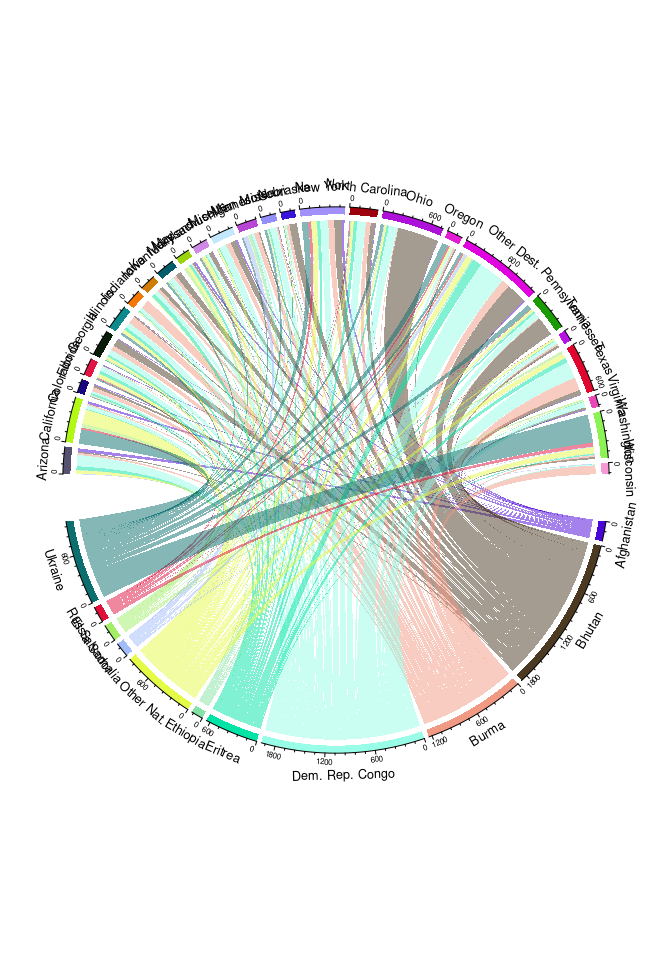
Gapminder Childhood Mortality Data
The gapminder package provides a subset of the data from the Gapminder web site. Additional data sets are available.
A data set on childhood mortality is available locally as a csv file or an Excel file.
The numbers represent number of deaths within the first five years per 1000 births.
Loading the data:
if (! file.exists("gapminder-under5mortality.xlsx"))
download.file("http://homepage.stat.uiowa.edu/~luke/data/gapminder-under5mortality.xlsx", "gapminder-under5mortality.xlsx")
gcm <- read_excel("gapminder-under5mortality.xlsx")
names(gcm)[1]
## [1] "Under five mortality"
names(gcm)[1] <- "country"A tidy version is useful for working with ggplot.
tgcm <- gather(gcm, year, u5mort, -1)
head(tgcm)
## # A tibble: 6 × 3
## country year u5mort
## <chr> <chr> <dbl>
## 1 Abkhazia 1800.0 NA
## 2 Afghanistan 1800.0 469.
## 3 Akrotiri and Dhekelia 1800.0 NA
## 4 Albania 1800.0 375.
## 5 Algeria 1800.0 460.
## 6 American Samoa 1800.0 NA
tgcm <- mutate(tgcm, year = as.numeric(year))
head(tgcm)
## # A tibble: 6 × 3
## country year u5mort
## <chr> <dbl> <dbl>
## 1 Abkhazia 1800 NA
## 2 Afghanistan 1800 469.
## 3 Akrotiri and Dhekelia 1800 NA
## 4 Albania 1800 375.
## 5 Algeria 1800 460.
## 6 American Samoa 1800 NAA multiple time series version may also be useful.
gcmts <- ts(t(gcm[-1]), start = 1800)
colnames(gcmts) <- gcm$countrySome explorations:
p <- ggplot(tgcm) + geom_line(aes(year, u5mort, group = country), alpha = 0.3)
p
## Warning: Removed 18644 rows containing missing values or values outside the scale range
## (`geom_line()`).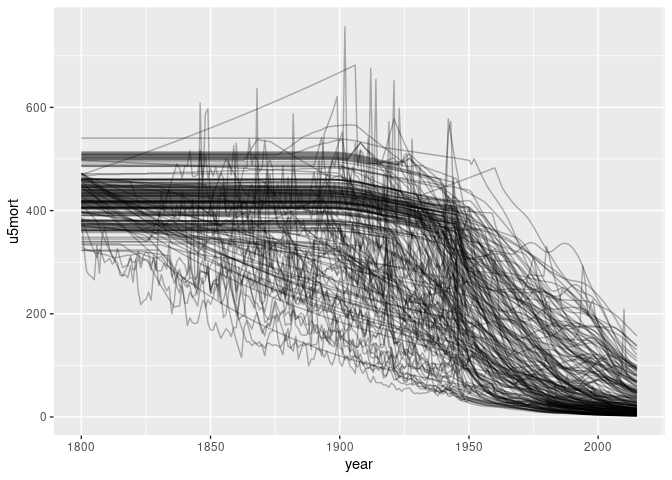
plotly::ggplotly(p)
xyplot(gcmts, superpose = TRUE, auto.key = FALSE)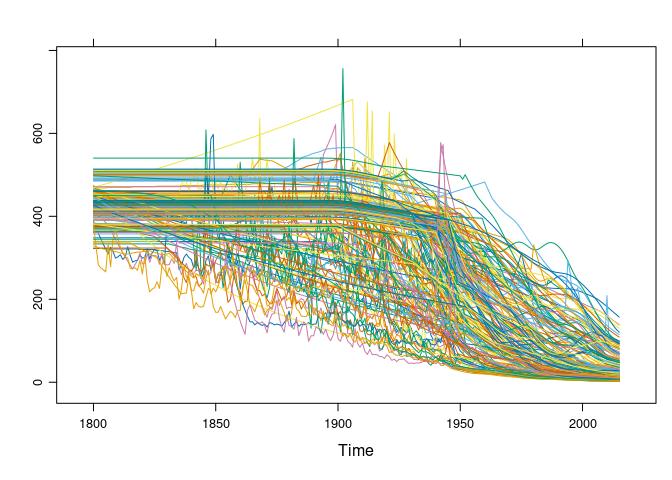
xyplot(gcmts, superpose = TRUE, auto.key = FALSE, col = "black")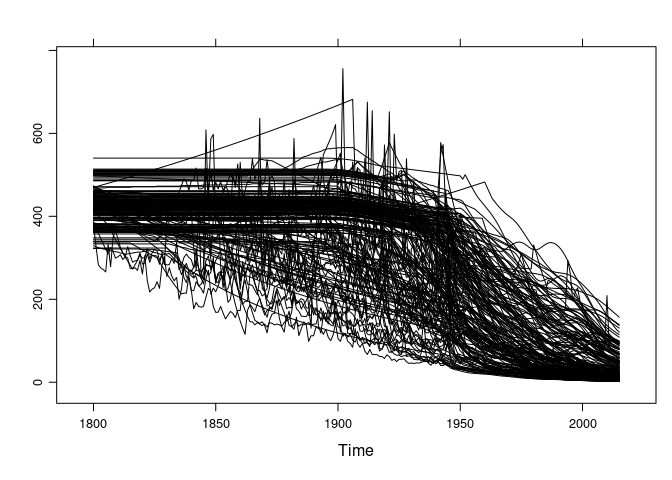
Examining the missing values:
anyNA(gcmts)
## [1] TRUE
sum(is.na(gcmts))
## [1] 18644
length(gcmts)
## [1] 59400
image(time(gcmts), seq_len(ncol(gcmts)), is.na(gcmts), ylab = "series")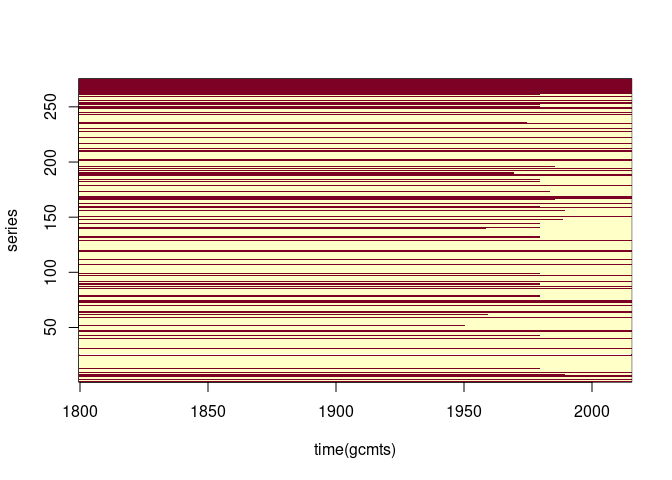
Re-ordering by the number of missing values may be useful:
naord <- order(apply(gcmts, 2, function(x) sum(is.na(x))))
image(time(gcmts), seq_len(ncol(gcmts)), is.na(gcmts)[, naord], ylab = "series")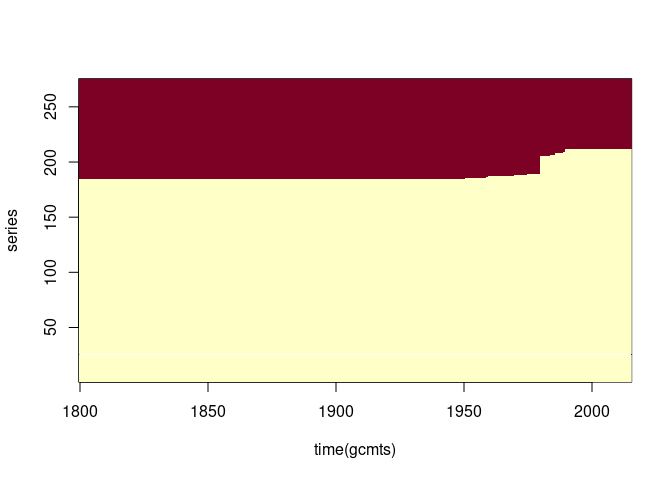
Some series look like the early values are estimates. Identifying and removing the ones that are constant between 1800 and 1850:
gcmtsNoNA <- gcmts[, ! apply(gcmts, 2, anyNA)]
i1850 <- which(time(gcmts) == 1850)
early_sd <- apply(gcmtsNoNA[1 : i1850, ], 2, sd)
filled_in <- names(which(early_sd == 0))
ggplot(filter(tgcm, ! country %in% filled_in)) +
geom_line(aes(year, u5mort, group = country), alpha = 0.3, na.rm = TRUE)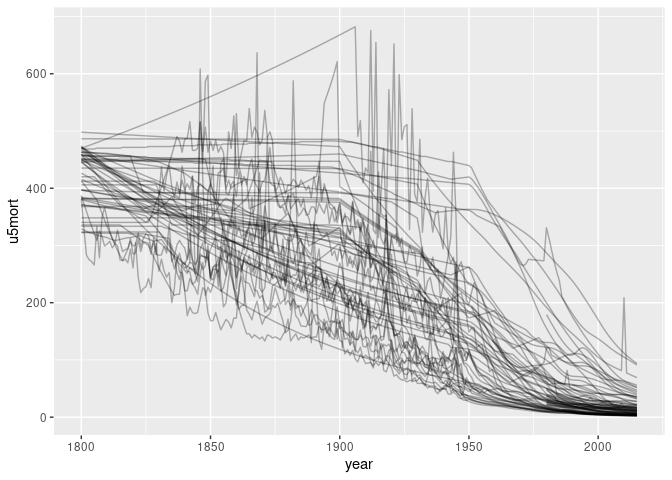
To show mortality rates by continent we can use the continent identification in the gapminder data set of the gapminder package.
There are a number of countries in the new data set not in gapminder
library(gapminder)
dim(gcm)
## [1] 275 217
length(unique(gapminder$country))
## [1] 142
head(setdiff(gcm$country, gapminder$country))
## [1] "Abkhazia" "Akrotiri and Dhekelia" "American Samoa"
## [4] "Andorra" "Anguilla" "Antigua and Barbuda"There are several options for adding the continent information from gapminder to one of our data frames:
look-up using match
a left join or inner join using
left_joina left join or inner join using
merge.
Using match, look up the index of the first row in gapminder for the country; then look up the corresponding continent:
cidx <- match(tgcm$country, gapminder$country)
head(cidx)
## [1] NA 1 NA 13 25 NA
cont <- gapminder$continent[cidx]
head(cont)
## [1] <NA> Asia <NA> Europe Africa <NA>
## Levels: Africa Americas Asia Europe Oceania
d1 <- mutate(tgcm, continent = cont)Our left table will map country to continent, with one row per country:
country_continent <- unique(select(gapminder, country, continent))
head(country_continent)
## # A tibble: 6 × 2
## country continent
## <fct> <fct>
## 1 Afghanistan Asia
## 2 Albania Europe
## 3 Algeria Africa
## 4 Angola Africa
## 5 Argentina Americas
## 6 Australia OceaniaThe left join using left_join:
d2 <- left_join(tgcm, country_continent)
## Joining with `by = join_by(country)`Joins in general may not produce the ordering you want; you need to check or make sure by reordering:
d1 <- arrange(d1, year, country)
d2 <- arrange(d2, year, country)
identical(unclass(d1), unclass(d2))
## [1] TRUEThe left join using merge:
d3 <- merge(tgcm, country_continent, all.x = TRUE)
d3 <- arrange(d3, year, country)
identical(d1, d3)
## [1] FALSEMapping continent to color:
ggplot(d3) +
geom_line(aes(year, u5mort, group = country, color = continent),
alpha = 0.7, na.rm = TRUE)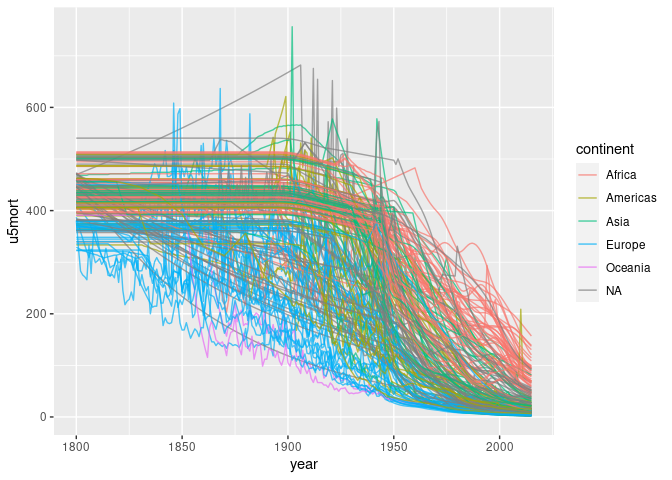
To compare child mortality to GDP per capita for the years covered by the gapminder data frame we need to merge the mortality data into gapminder.
Some country names in gapminder do not appear in the child mortality data:
setdiff(gapminder$country, tgcm$country)
## [1] "Korea, Dem. Rep." "Korea, Rep." "Yemen, Rep."
grep("Korea", unique(tgcm$country), value = TRUE)
## [1] "North Korea" "South Korea"
## [3] "United Korea (former)\n"
grep("Yemen", unique(tgcm$country), value = TRUE)
## [1] "North Yemen (former)" "South Yemen (former)" "Yemen"Change the names in tgcm`` to match the names ingapminder`:
ink <- which(tgcm$country == "North Korea")
isk <- which(tgcm$country == "South Korea")
iym <- which(tgcm$country == "Yemen")
tgcm$country[ink] <- "Korea, Dem. Rep."
tgcm$country[isk] <- "Korea, Rep."
tgcm$country[iym] <- "Yemen, Rep."Now left join the mortality data with gapminder; the key is the combination of country and year:
Some plots:
gm <- left_join(gapminder, tgcm)
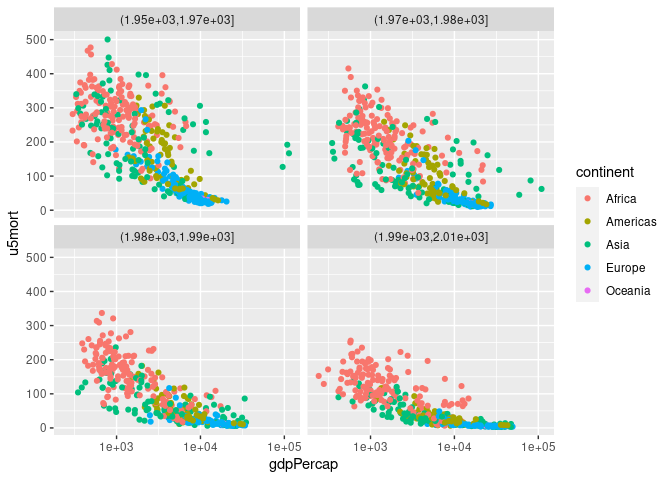
## Joining with `by = join_by(country, year)`p <- ggplot(gm, aes(gdpPercap, u5mort, color = continent)) + geom_point()
p
## Warning: Removed 30 rows containing missing values or values outside the scale range
## (`geom_point()`).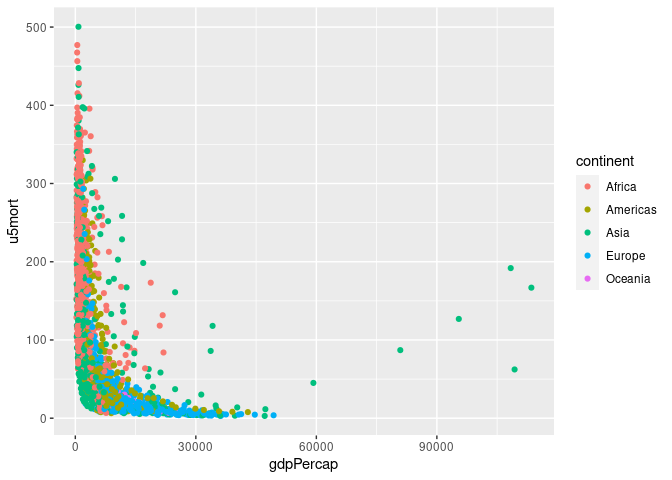
p + scale_x_log10()
## Warning: Removed 30 rows containing missing values or values outside the scale range
## (`geom_point()`).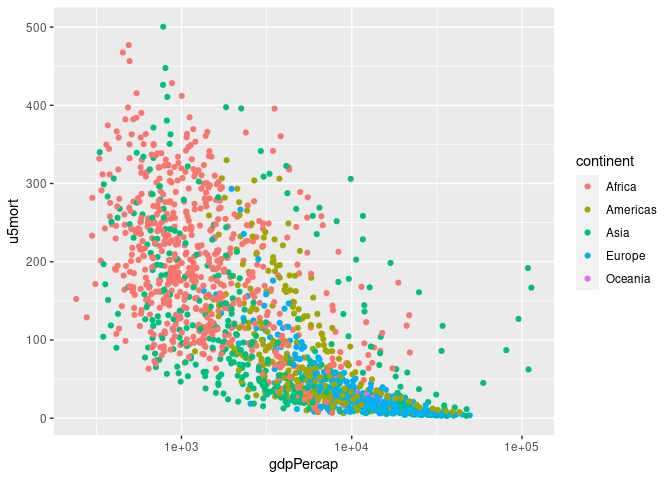
p + scale_x_log10() + facet_wrap(~ cut(year, 4))
## Warning: Removed 30 rows containing missing values or values outside the scale range
## (`geom_point()`).